Bahar Yilmazel, Yanhui Hu, Frederic Sigoillot, Jennifer A Smith, Caroline E Shamu, Norbert Perrimon, and Stephanie E Mohr 14 "Online GESS prediction of miRNAlike offtarget effects in largescale RNAi screen data by seed region analysis" BMC Bioinformatics, 15, Pp 192Basepairing of the socalled miRNA "seed" region with mRNAs identifies many thousands of putative targets Evaluating the strength of the resulting mRNA repression remains challenging, but is essential for a biologically informative ranking of potential miRNA targetsCanonically, miRNA targeting is reliant on base pairing of the seed region, nucleotides 2–7, of the miRNA to sites in mRNA 3′ untranslated regions Recently, the 3′ half of the miRNA has gained attention for newly appreciated roles in regulating target specificity and regulation
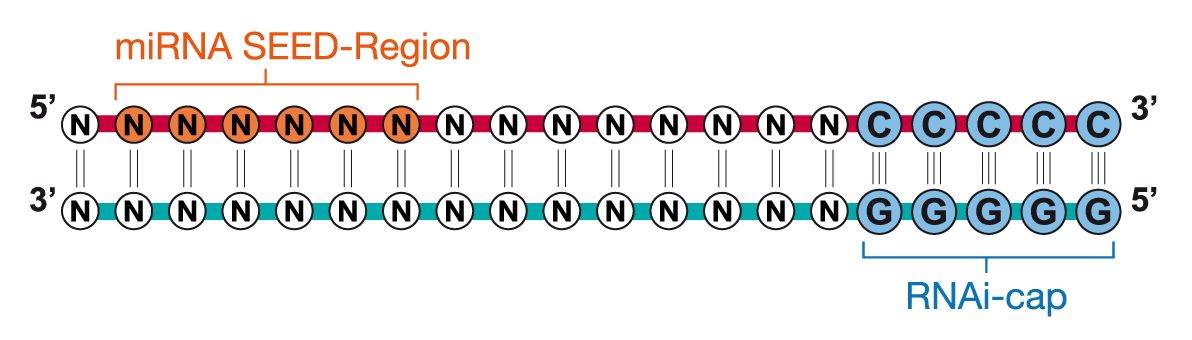
Riboxx Rna Technologies Benefits Of Rnai Cap For Mirna
Seed region of mirna
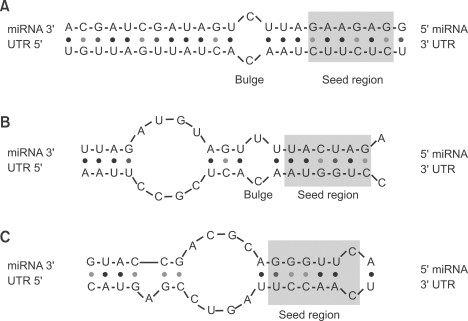
Seed region of mirna-A mismatch tolerance test assay, based on pools of transgenic strains, revealed that target hybridization to nucleotides of the seed region, at the 5′ end of an miRNA, was sufficient to induce moderate repression of expression In contrast, pairing to the 3′ region of the miRNA was not critical for silencingThe seed sequence or seed region is a conserved heptametrical sequence which is mostly situated at positions 27 from the miRNA 5´end Even though base pairing of miRNA and its target mRNA does not match perfect, the "seed sequence" has to be perfectly complementary
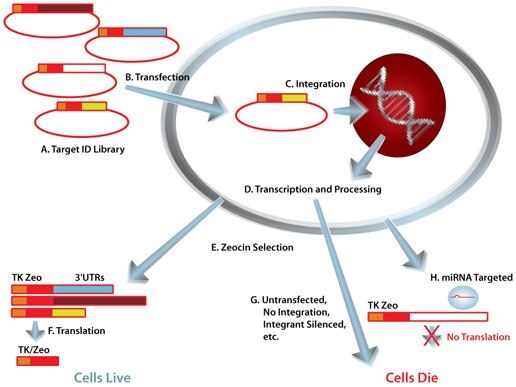



Mission Target Id Library For Human Mirna Target Id And Discovery
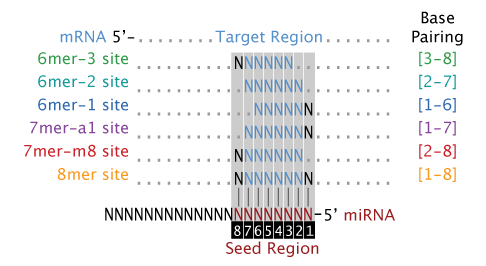
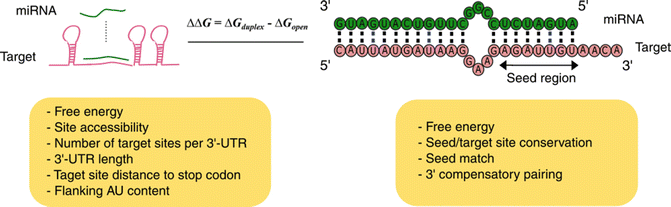
Defined as six or seven nucleotides between the nucleotides 27 or 28 of the miRNA 5' end that are responsible for mRNA binding 11(Figure (Figure11C) Seed Pairing Is Enriched in miRNATarget Sites The miRNA sequence can be separated into five functional domains that affect miRNAtarget recognition 5′ anchor (nt 1), seed sequence (nts 2–8), central region (nts 9–12), 3′ supplementary region (nts 13–16), and 3′ tail (nts 17–22) (Wee et al, 12)Online GESS prediction of miRNAlike offtarget effects in largescale RNAi screen data by seed region analysis Bahar Yilmazel1, Yanhui Hu1, Frederic Sigoillot2, Jennifer A Smith3, Caroline E Shamu3, Norbert Perrimon1,4 and Stephanie E Mohr1* Abstract Background RNA interference (RNAi) is an effective and important tool used to study gene
MicroRNAs (miRNAs) bind to mRNAs and target them for translational inhibition or transcriptional degradation It is thought that most miRNAmRNA interactions involve the seed region at the 5′ end of the miRNA The importance of seed sites is supported by experimental evidence, although there is growing interest in interactions mediated by the central region of the miRNA To characterize the basepairing patterns of miRNAtarget interaction, we searched for overrepresented sequence elements in all the targets discovered for each miRNA For most of the top expressed miRNAs, highly enriched sequence motifs emerged as complementary to the extended miRNA seed region (nucleotides 1–8 of miRNA) (Fig 4A and S7 Fig) The impact of miRNA seed types on target downregulation Previous studies have identified several major types of canonical miRNA target sites, including those matching to the 6mer, 7mer, or 8mer miRNA seed sequences (Table 2)Sequence conservation analysis suggested that target sites pairing to longer miRNA seeds are more conserved across species and thus are
Results from luciferasereporter assays and global expression data confirmed previous observations that the siRNA seed region is the primary determinant for offtarget gene recognition and bindingMicroRNAs (miRNAs) are key regulators of sequencespecific gene silencing However, crucial factors that determine the efficacy of miRNAmediated target gene silencing are poorly understood Here we mathematized basepairing stability and showed that miRNAs with an unstable 5′ terminal duplex and stable seedtarget duplex exhibit strong silencing activityAs a result, we identified 48 SNPs in human miRNA seed regions and thousands of SNPs in 3' untranslated regions with the potential to either disturb or create miRNAtarget interactions Furthermore, we experimentally confirmed seven lossoffunction SNPs and one gainoffunction SNP by luciferase assay
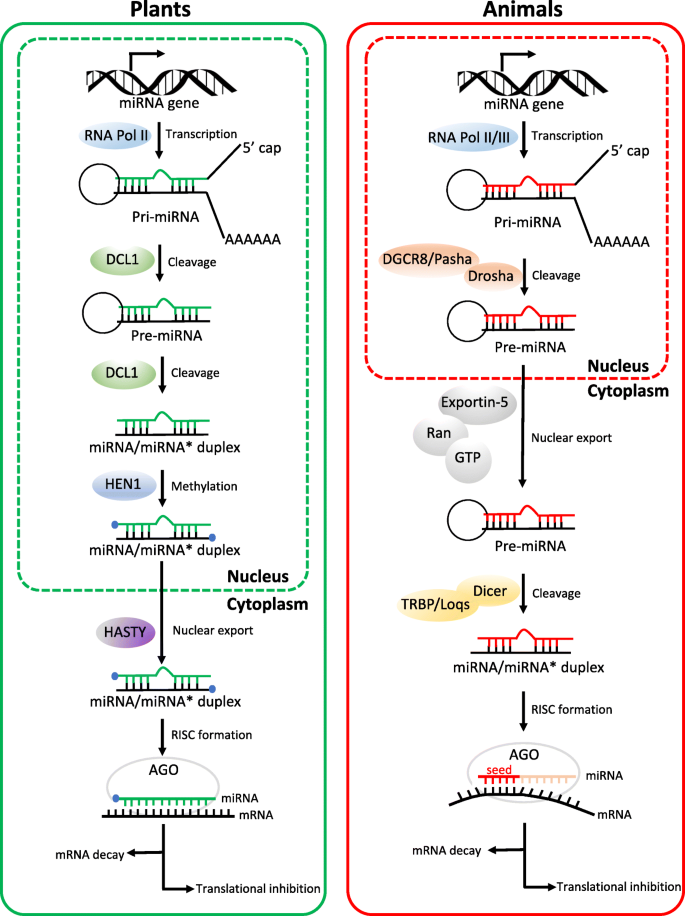



Micrornas From Plants To Animals Do They Define A New Messenger For Communication Nutrition Metabolism Full Text




Correlation Between Mirna Targeted Gene Promoter F1000research
In human miRNA seed regions A total of 1879 SNPs were mapped to 1226 human miRNA seed regions We found that miRNAs with SNPs in their seed region are significantly enriched in miRNA clusters We also found that SNPs in clustered miRNA seed regions have a lower functional effect than have SNPs in nonclustered miRNA seed regions Additionally, weIn molecular genetics, the three prime untranslated region (3′UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codonThe 3′UTR often contains regulatory regions that posttranscriptionally influence gene expression During gene expression, an mRNA molecule is transcribed from the DNA sequence and is later translated into a proteinSOFTWARE Open Access Online GESS prediction of miRNAlike offtarget effects in largescale RNAi screen data by seed region analysis Bahar Yilmazel1, Yanhui Hu1, Frederic Sigoillot2, Jennifer A Smith3, Caroline E Shamu3, Norbert Perrimon1,4 and Stephanie E Mohr1* Abstract



Mirvestigator Framework Detect The Mirnas Driving Co Expression Signatures
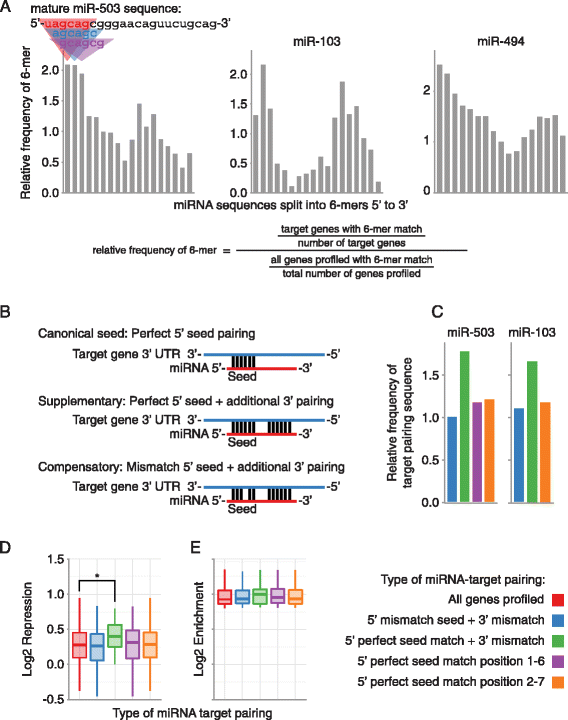



Mir 503 Represses Human Cell Proliferation And Directly Targets The Oncogene Ddhd2 By Non Canonical Target Pairing Bmc Genomics Full Text
Reveal that miRNA containing inosine in the seed region exhibits a different degree of silencing efficiency compared to the corresponding miRNA with guanosine at the same position The difference in basepairing stability, deduced by melting temperature measurements, between seedtarget duplexes containing either CG or IC pairs may account for Bases 2–8 (seed region) from the 5′end of the mature miRNA are critical determinants of target complementarity 22 Premature forms of a miRNAMouse let7 miRNA populations exhibit RNA editing that is constrained in the 5'seed/ cleavage/anchor regions and stabilize predicted mmulet7amRNA duplexes 11



Journal Hep Com Cn Pac En Article Downloadarticlefile Do Attachtype Pdf Id 1345




Stability Of Mirna 5 Terminal And Seed Regions Is Correlated With Experimentally Observed Mirna Mediated Silencing Efficacy Topic Of Research Paper In Biological Sciences Download Scholarly Article Pdf And Read For Free On
After the siRNA seed region anneals, the catalytic RNase H domain of Argonaute then subjects perfectly complementary mRNA sequences 10 nucleotides from the 5' end of the incorporated siRNA strand to nucleolytic degradation, resulting inTargetScan does its own classification of miRNAs into families, based on identical seed region, so miRNA family classification and, even more likely, nomenclature can differ between these databases In TargetScan 5 Custom, nonconserved target sites are not shown; Part of a miRNA is a 2–8base pair long seed region in their 5′ end The interaction happens between these seed regions and complementary "seed matches" on the target sites of the mRNAs 7
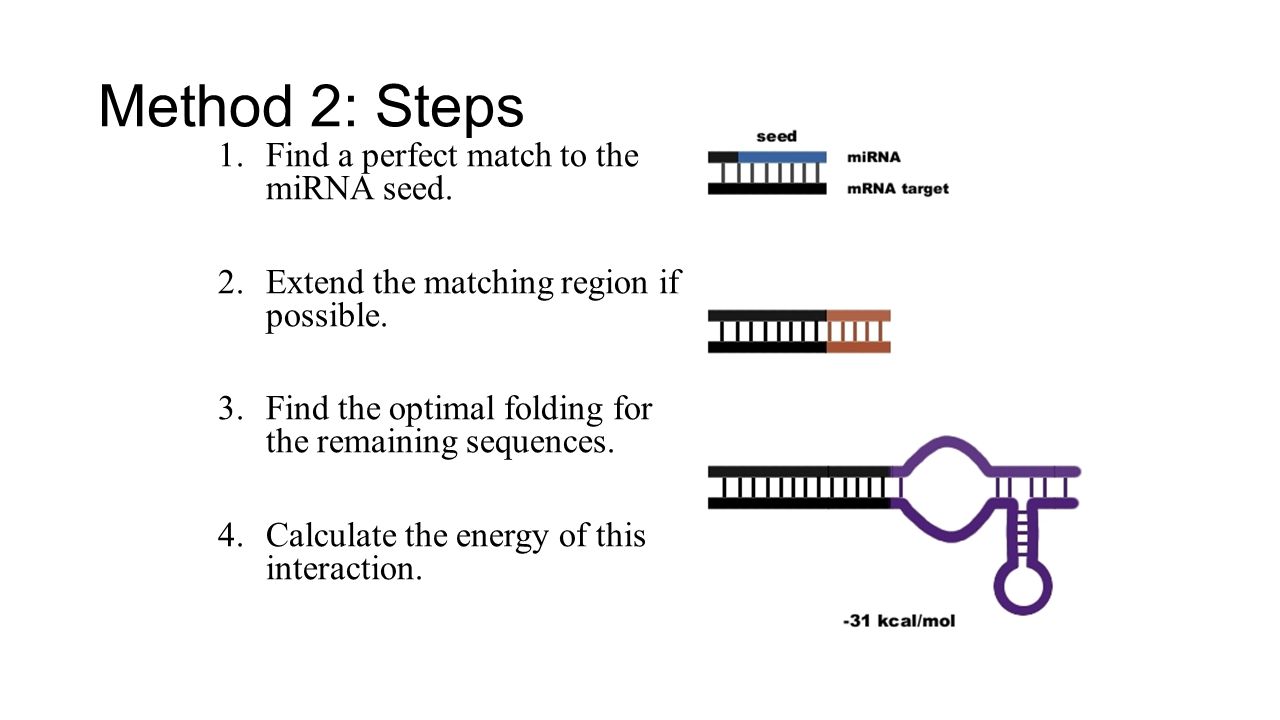



Microrna Computational Prediction And Analysis Ppt Download




Prediction Of Human Mirnas Using Tissue Selective Motifs In 3 Utrs Pnas
For the 1226 human miRNAs with SNPs in their seed region, 314 (256%) of them are located in miRNA clusters, whereas among the 1587 human miRNAs without SNPs in their seed region, only 3 (2%) of them are located in miRNA clusters (P = 606 x 10sup4, chi square test) (Table S2) miRNAs from the same cluster have the tendency toTargetScan predicts biological targets of miRNAs by searching for the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA (Lewis et al, 05)As an option, predictions with only poorly conserved sites are also providedThe most critical region for complementarity is the seed region (nucleotides 2–7 from the 5'terminus of the miRNA) 4,7 Aside from the seed region, other criteria have been established that can enhance miRNAmediated repression including complementarity at position 8 and the presence of an adenosine residue opposite the first miRNA




Snps In Microrna Target Sites And Their Potential Role In Human Disease Open Biology




Mirna Targeting Growing Beyond The Seed Trends In Genetics
MiRNA seed region is more critical than the 3′ region for target recognition in A thaliana (Mallory et al, 04) Moreover, plant and algal small RNAs also induce translational repression of perfectly complementary target mRNAs without, or with only minimal, transcript destabiliTargetScan predicts biological targets of miRNAs by searching for the presence of 8mer, 7mer, and 6mer sites that match the seed region of each miRNA As an option, only conserved sites are predicted Also identified are sites with mismatches in the seed region that are compensated by conserved 3' pairing and centered sitesThe latest version of this resource was released in August 15 miRSearch



Ynupjjdamdxssm




Pairing Beyond The Seed Supports Microrna Targeting Specificity Sciencedirect
SCIENTIFIC REPORTS srep 1 wwwnaturecomscientificreports Long noncoding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes Alireza Ahadi1,2, Samuel Brennan3, Paul J Kennedy2,4, Gyorgy Hutvagner2 & Nham Tran2,5 Long noncoding RNAs (lncRNAs) form the largest transcript class in the human transcriptomeTargetScan is a target prediciton tool that predicts biological targets of miRNAs by searching for the presence of conserved 8mer and 7mer sites that match the seed region of each miRNA The target prediction software is frequently updated;I microRNA si appaiano all'mRNA target attraverso i nucleotidi 28 del miRNA (seed region) e il sito complementare della regione 3'UTR dell'mRNA bersaglio PDF PDF
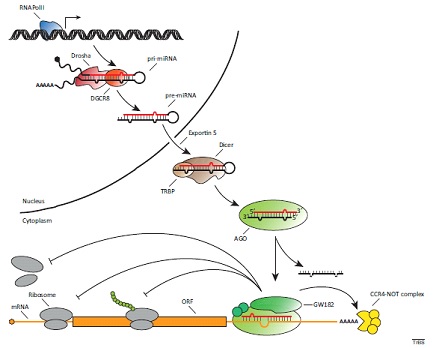



Identification Of Non Coding Cellular Rna As New Antiviral Targets
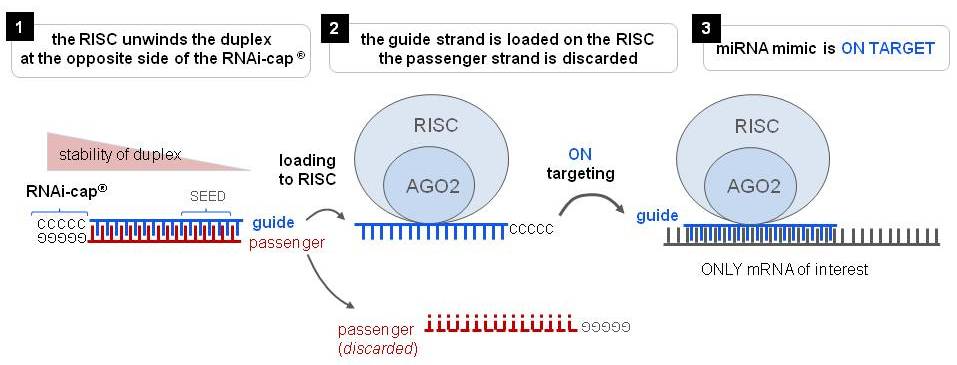



Riboxx Rna Technologies Benefits Of Rnai Cap For Mirna
A mismatch tolerance test assay, based on pools of transgenic strains, revealed that target hybridization to nucleotides of the seed region, at the 5′ end of an miRNA, was sufficient to induce moderate repression of expression In contrast, pairing to the 3′ region of the miRNA was not critical for silencingComplementarity to an miRNA Seed Region Is Sufficient to Induce Moderate Repression of a Target Transcript in the Unicellular Green Alga Chlamydomonas reinhardtii Tomohito Yamasaki,1 Adam Voshall,2 EunJeong Kim,2 Etsuko Moriyama,2 Heriberto Cerutti,2 and Takeshi Ohama1 1Specifically, seedbased canonical target recognition was dependent on the GC content of the miRNA seed For miRNAs with low GC content of the seed region, noncanonical targeting was the dominant mechanism for target recognition



Endogenous Transcripts Control Mirna Levels And Activity In Mammalian Cells By Target Directed Mirna Degradation Nature Communications




Snps In Microrna Target Sites And Their Potential Role In Human Disease Open Biology
For the 1226 human miRNAs with SNPs in their seed region, 314 (256%) of them are located in miRNA clusters, whereas among the 1587 human miRNAs without SNPs in their seed region, only 3 (2%) of them are located in miRNA clusters (P = 606 × 10 −4, χ 2 test) (Table S2) miRNAs from the same cluster have the tendency to regulate the One class of pattern consists of perfect WatsonCrick binding at the 5'end of the miRNA This region is known as "seedregion" and found at the 27 base of the miRNA This region is able to suppress the target mRNAs without having a complete base pairing at Further studies revealed that the pairing of miRNA position 8 was dispensable in many cases, indicating that the seed region can be as
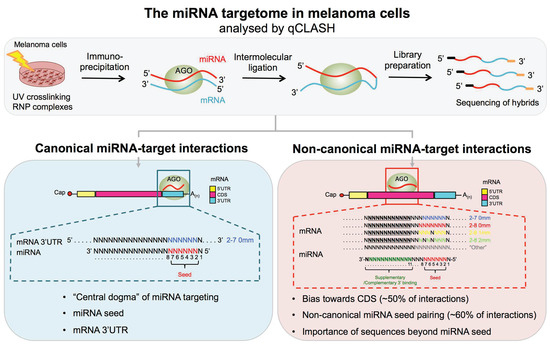



Cancers Free Full Text Cross Linking Ligation And Sequencing Of Hybrids Qclash Reveals An Unpredicted Mirna Targetome In Melanoma Cells Html




Rna Interference The Molecules That Govern Genetic Control Young Scientists Journal
However, this information is available for annotated miRNAs This mature miRNA SNP (miRSNP) is located within the "seed region";The seedregion are rare and comprise less than 1% of the miRNA The role of miRNAs in processes such as maturation of the related SNPs 22 mammalian kidney was recently established by the podocyte MicroRNA associated single nucleotide polymorphisms specific inactivation of Dicer, the RNAse III endonuclease (miRSNPs) found on miRNA target



Miraw A Deep Learning Based Approach To Predict Microrna Targets By Analyzing Whole Microrna Transcripts




Pairing Beyond The Seed Supports Microrna Targeting Specificity Sciencedirect
And some miRNA nucleotides are more important than others (8, 9) Specifically, pairing to the miRNA "seed region" (nt 2 to 7 or 2 to 8, from the 5′ end) is the most evolutionarily conserved feature of miRNA targets in animals (10–14) Crystal structures of human Argonaute proteins show nt 2 to 6 of the guide RNA bound Furthermore, we evaluated the silencing efficiencies of miRNAs with the same seven nucleotide compositions in their seed regions (A = 3, U =




Mapping The Human Mirna Interactome By Clash Reveals Frequent Noncanonical Binding Cell



Upcommons Upc Edu Bitstream Handle 2117 Pdf Sequence 1 Isallowed Y
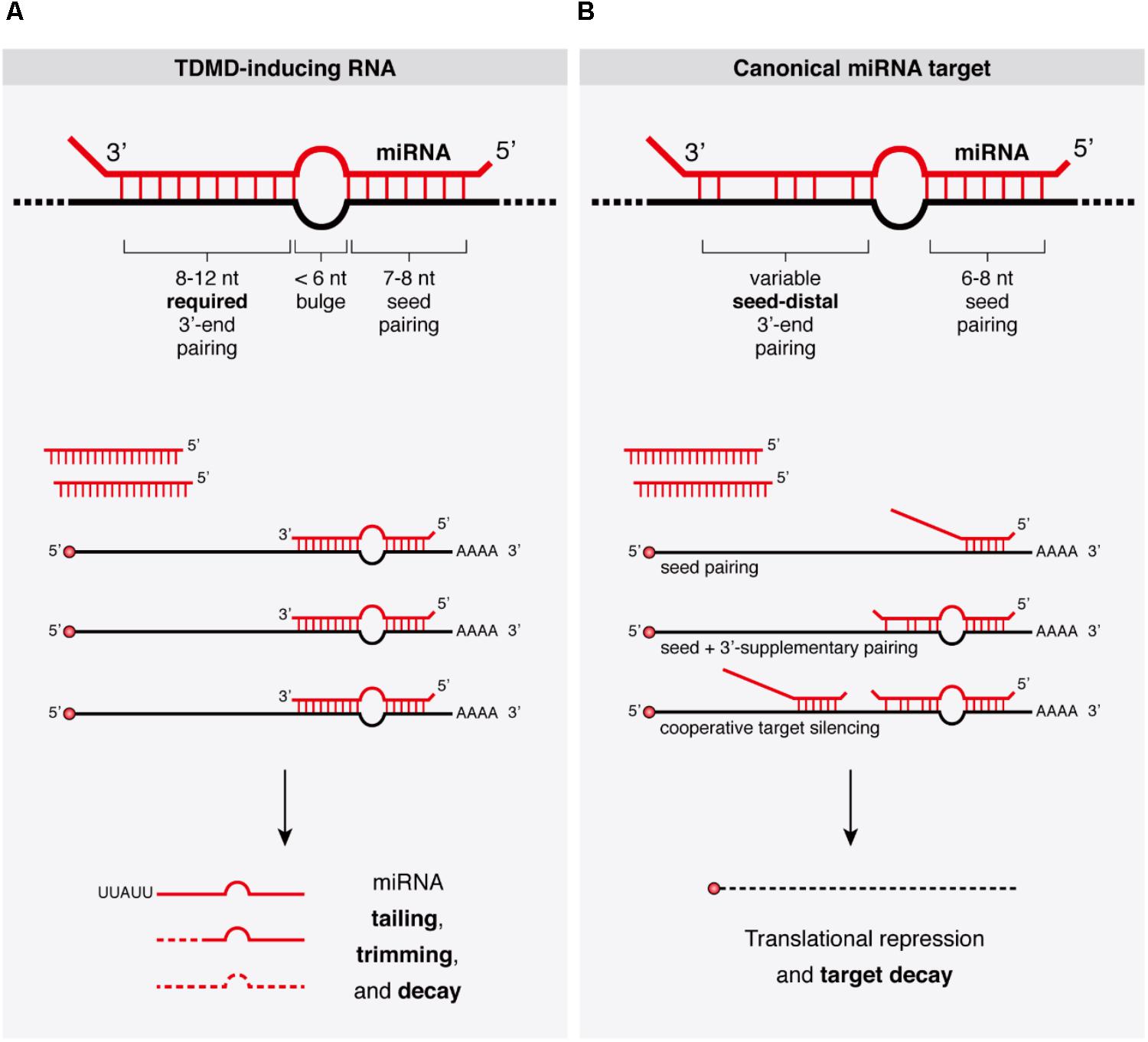



Frontiers Target Rnas Strike Back On Micrornas Genetics
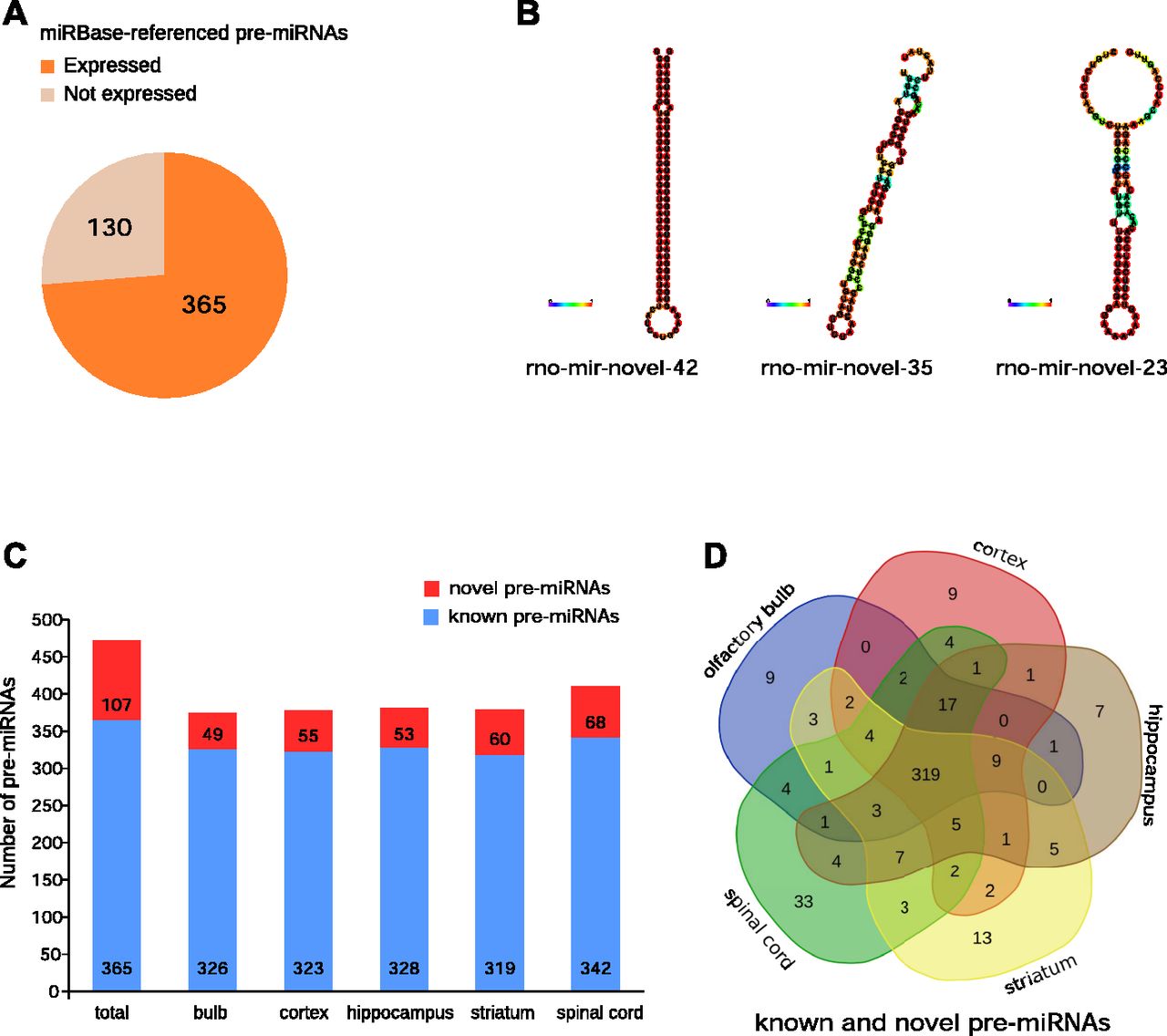



Small Rna Seq Reveals Novel Mirnas Shaping The Transcriptomic Identity Of Rat Brain Structures Life Science Alliance



Insect Micrornas From Molecular Mechanisms To Biological Roles Part 2




The Biochemical Basis Of Microrna Targeting Efficacy Biorxiv




Beyond The Seed Structural Basis For Supplementary Microrna Targeting By Human Argonaute2 The Embo Journal



Www Cell Com Cell Pdf S0092 8674 18 1 Pdf



1




Mismatches In The Mirna Proximal Seed Region Disrupt The Binding Download Scientific Diagram




Mirna Seed Types Nine Seed Types Are Categorized In Two Groups Download Scientific Diagram




Examples Of Mirna Target Interactions Pairing Schemes Open I




Introduction To Micrornas Ibiology



Mirepress Modelling Gene Expression Regulation By Microrna With Non Conventional Binding Sites Scientific Reports
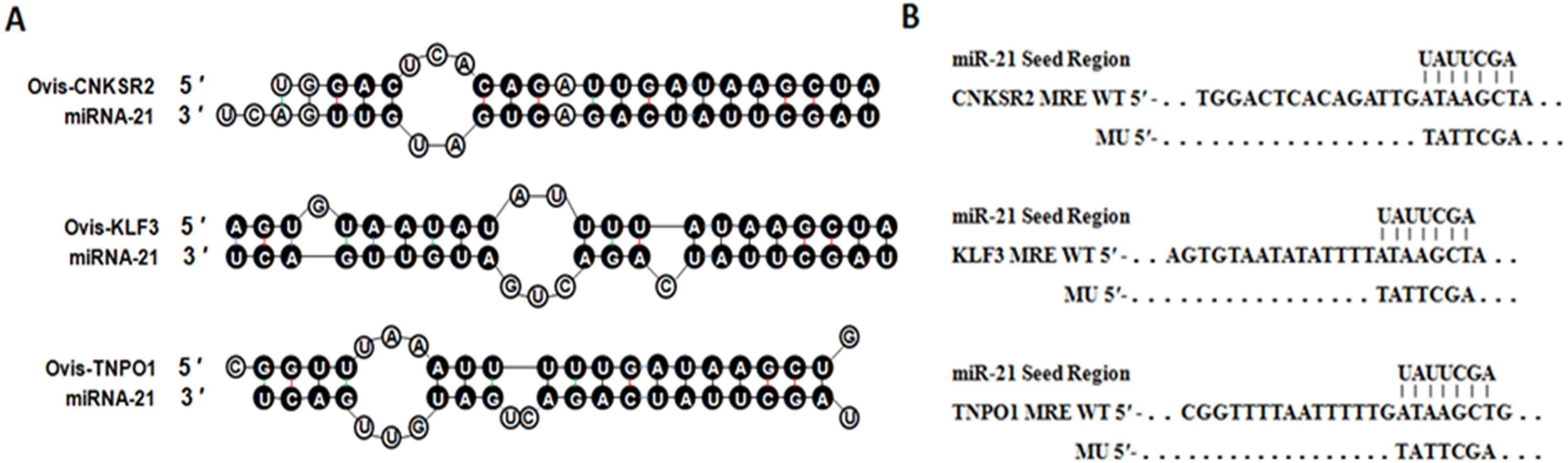



Identification Of Microrna 21 Target Genes Associated With Hair Follicle Development In Sheep Peerj




Riboxx Rna Technologies Benefits Of Rnai Cap For Mirna
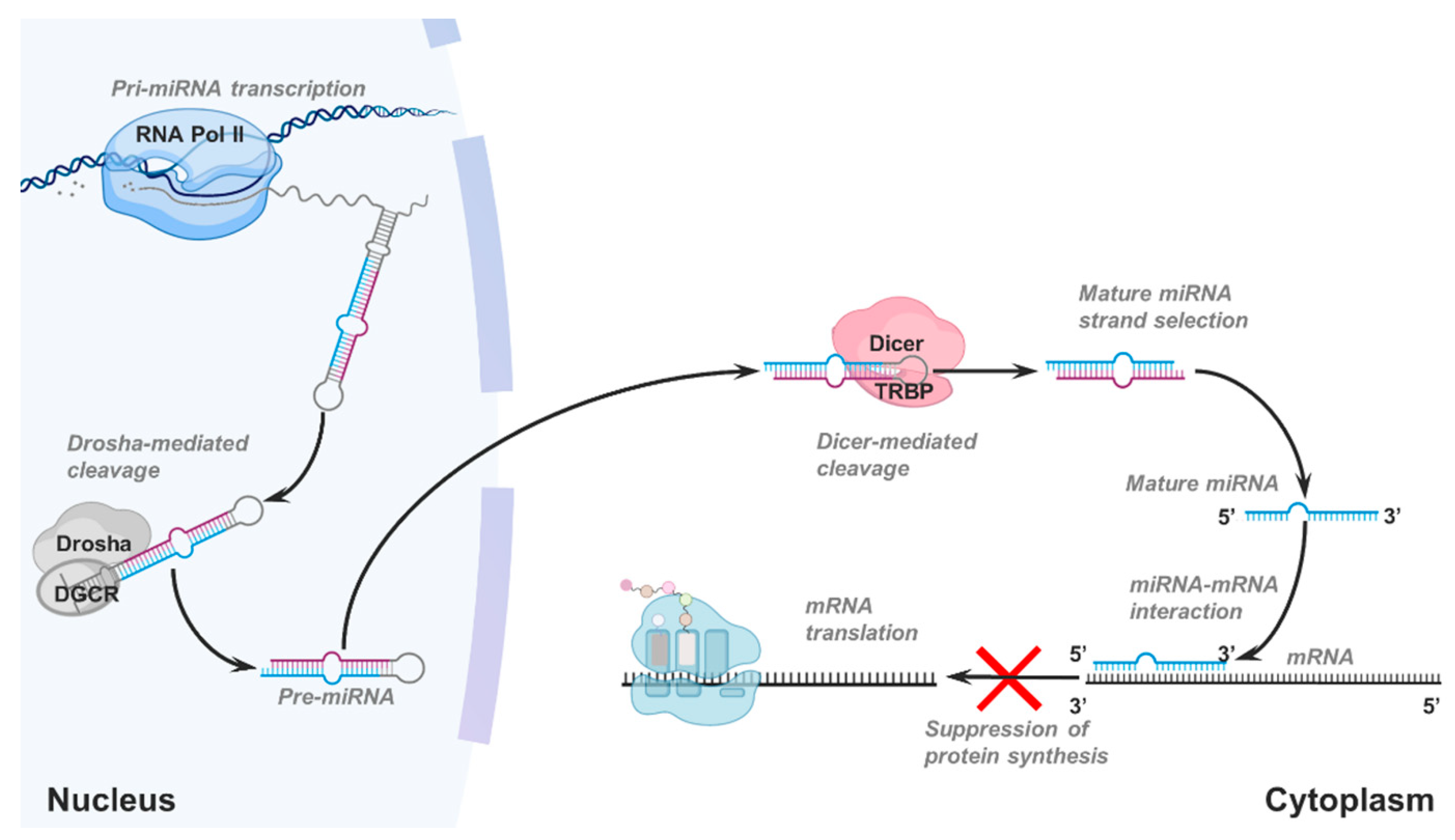



Cancers Free Full Text Detecting And Characterizing A To I Microrna Editing In Cancer Html




Beyond The Seed Structural Basis For Supplementary Microrna Targeting By Human Argonaute2 The Embo Journal



Stability Of Mirna 5 Terminal And Seed Regions Is Correlated With Experimentally Observed Mirna Mediated Silencing Efficacy Scientific Reports



Multimitar A Novel Multi Objective Optimization Based Mirna Target Prediction Method



Microrna Wikipedia




A Cartoon Showing The Site And Mechanism Of Mirna Targeting To Mrna Download Scientific Diagram




Rna Mrna Target Interaction Schematic Overview Of A Mirna Interaction Download Scientific Diagram




Crispr Screening Strategies For Microrna Target Identification Yang The Febs Journal Wiley Online Library
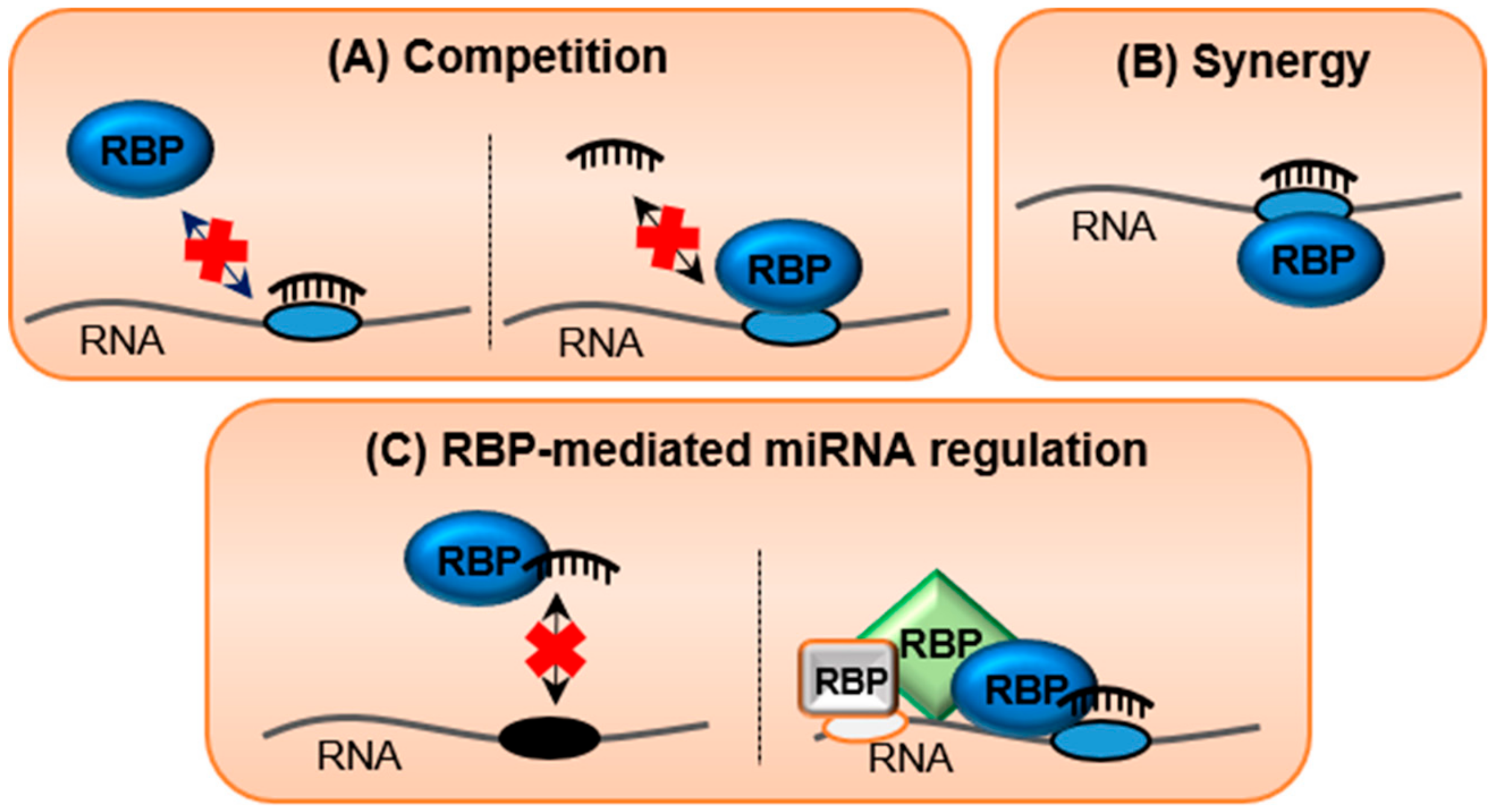



Cells Free Full Text The Butterfly Effect Of Rna Alterations On Transcriptomic Equilibrium Html




Systematic Prediction Of The Impacts Of Mutations In Microrna Seed Sequences




Structural Differences Between Pri Mirna Paralogs Promote Alternative Drosha Cleavage And Expand Target Repertoires Sciencedirect



The Sirna Non Seed Region And Its Target Sequences Are Auxiliary Determinants Of Off Target Effects




Mapping The Human Mirna Interactome By Clash Reveals Frequent Noncanonical Binding Abstract Europe Pmc



New Insights Into The Function Of Mammalian Argonaute2




Small Rna Seq Reveals Novel Mirnas Shaping The Transcriptomic Identity Of Rat Brain Structures Life Science Alliance
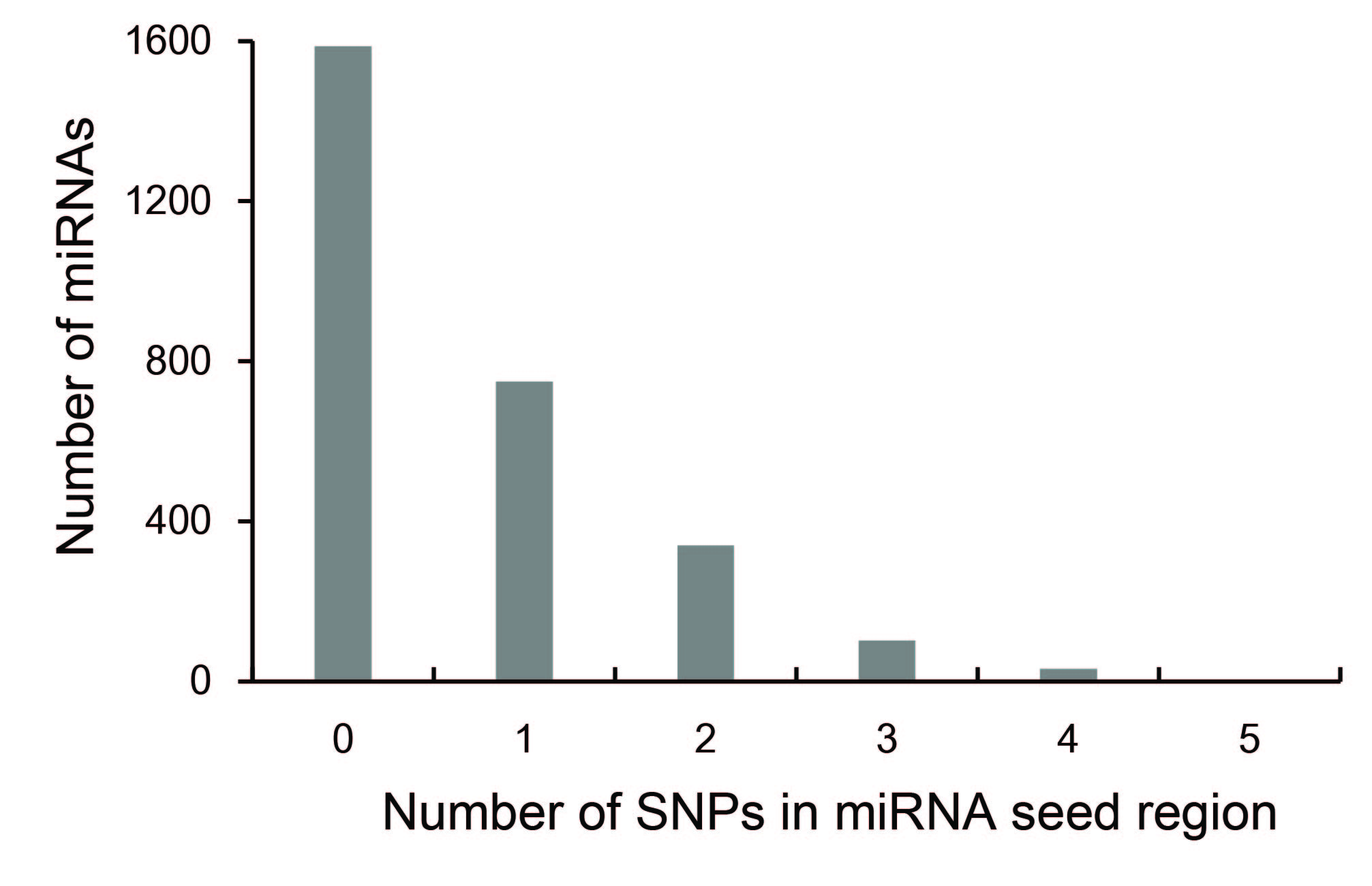



Clustering Pattern And Functional Effect Of Snps In Human Mirna Seed Regions



Silo Tips Download How To Measure Mirna Expression Matt Barter




Mirna Introduction Biogenesis Nomenclature And Experimental Workflow
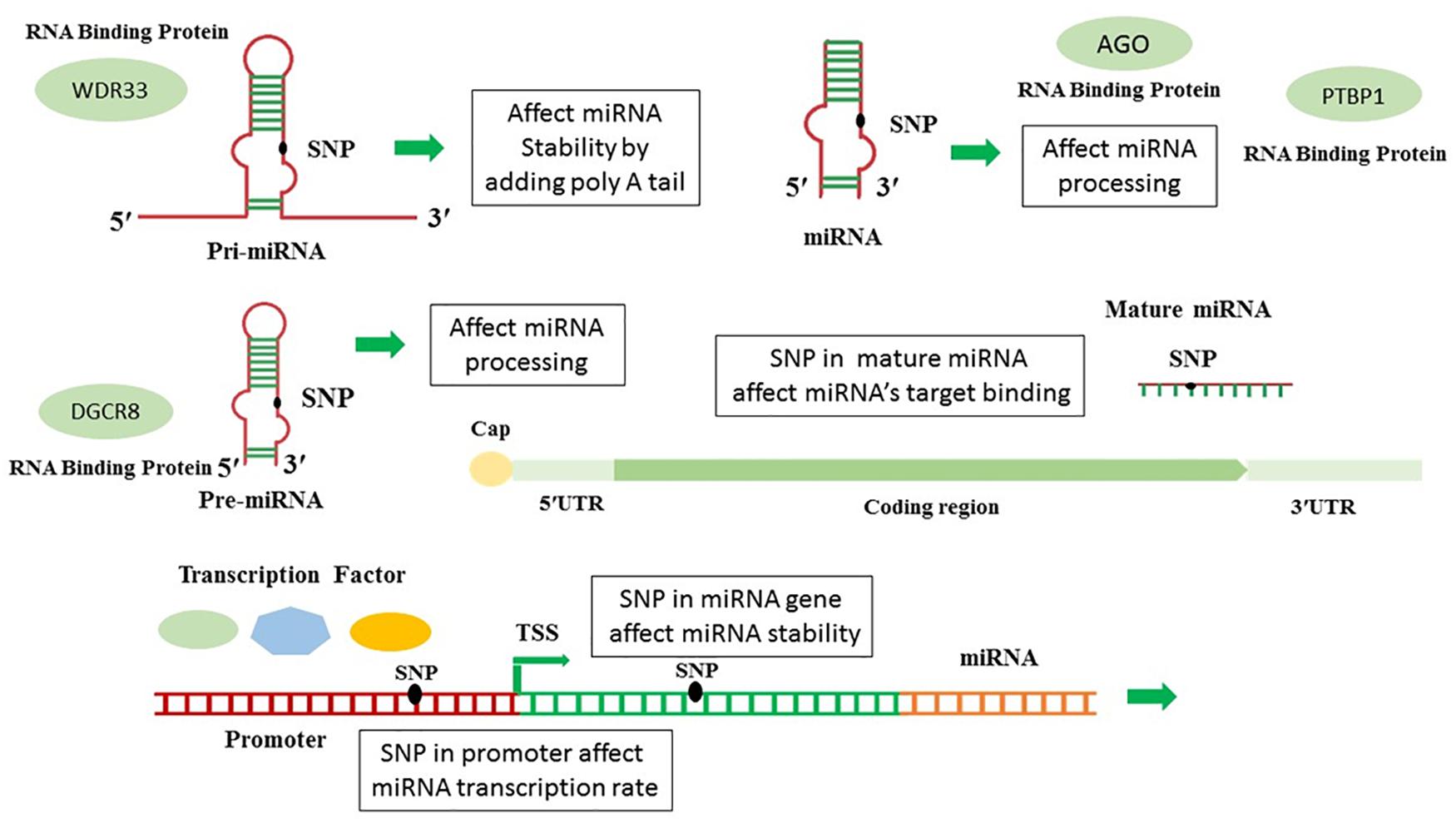



Frontiers In Silico Analysis Of Polymorphisms In Micrornas Deregulated In Alzheimer Disease Neuroscience




Helix 7 In Argonaute2 Shapes The Microrna Seed Region For Rapid Target Recognition The Embo Journal




Mapping The Human Mirna Interactome By Clash Reveals Frequent Noncanonical Binding Cell



Targetscan Non Canonical Sites




Mission Target Id Library For Human Mirna Target Id And Discovery



1




The Biochemical Basis Of Microrna Targeting Efficacy Science




Microrna Polymorphisms The Future Of Pharmacogenomics Molecular Epidemiology And Individualized Medicine Pharmacogenomics



1




Mirna Introduction Biogenesis Nomenclature And Experimental Workflow




Types Of Mirna Target Sites And Multiple Sites A Stringent Seed Download Scientific Diagram




Regulation Of Microrna Function In Animals Abstract Europe Pmc



Computational And Experimental Identification Of Tissue Specific Microrna Targets Springerlink



Got Target Computational Methods For Microrna Target Prediction And Their Extension Experimental Molecular Medicine




Microrna Wikipedia



Figure 1 Mir 106b Promotes Therapeutic Antibody Expression In Cho Cells By Targeting Deubiquitinase Cyld Springerlink




Sites Matching In The Mirna Seed Region Including All K Mer 8mer Download Scientific Diagram




Mirna Therapeutics A New Class Of Drugs With Potential Therapeutic Applications In The Heart Future Medicinal Chemistry
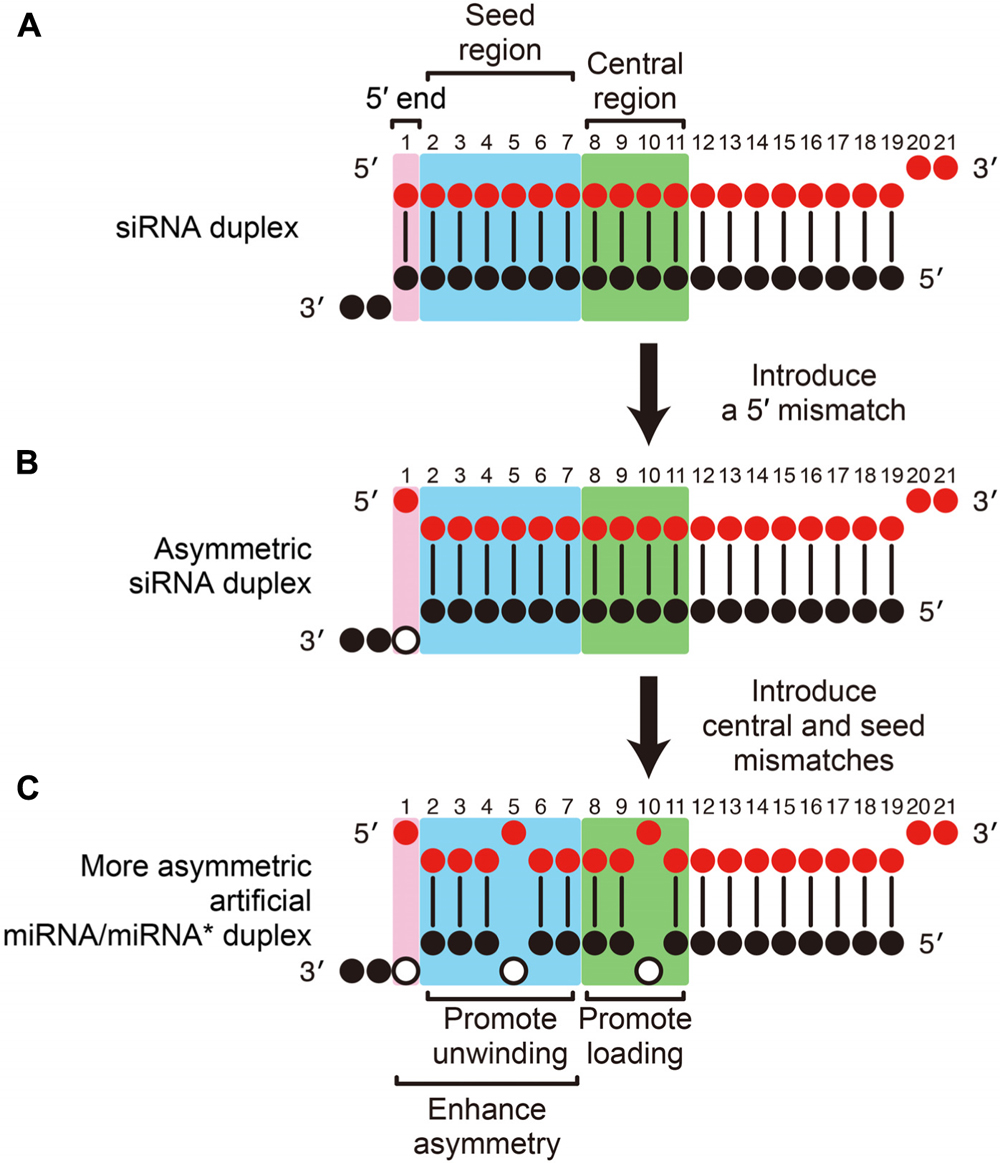



Frontiers Mirna Like Duplexes As Rnai Triggers With Improved Specificity Genetics



1




Tools For Sequence Based Mirna Ta Preview Related Info Mendeley



Pdfs Semanticscholar Org 5ff9 Cdd7524c67dbfcb3fa2 Pdf
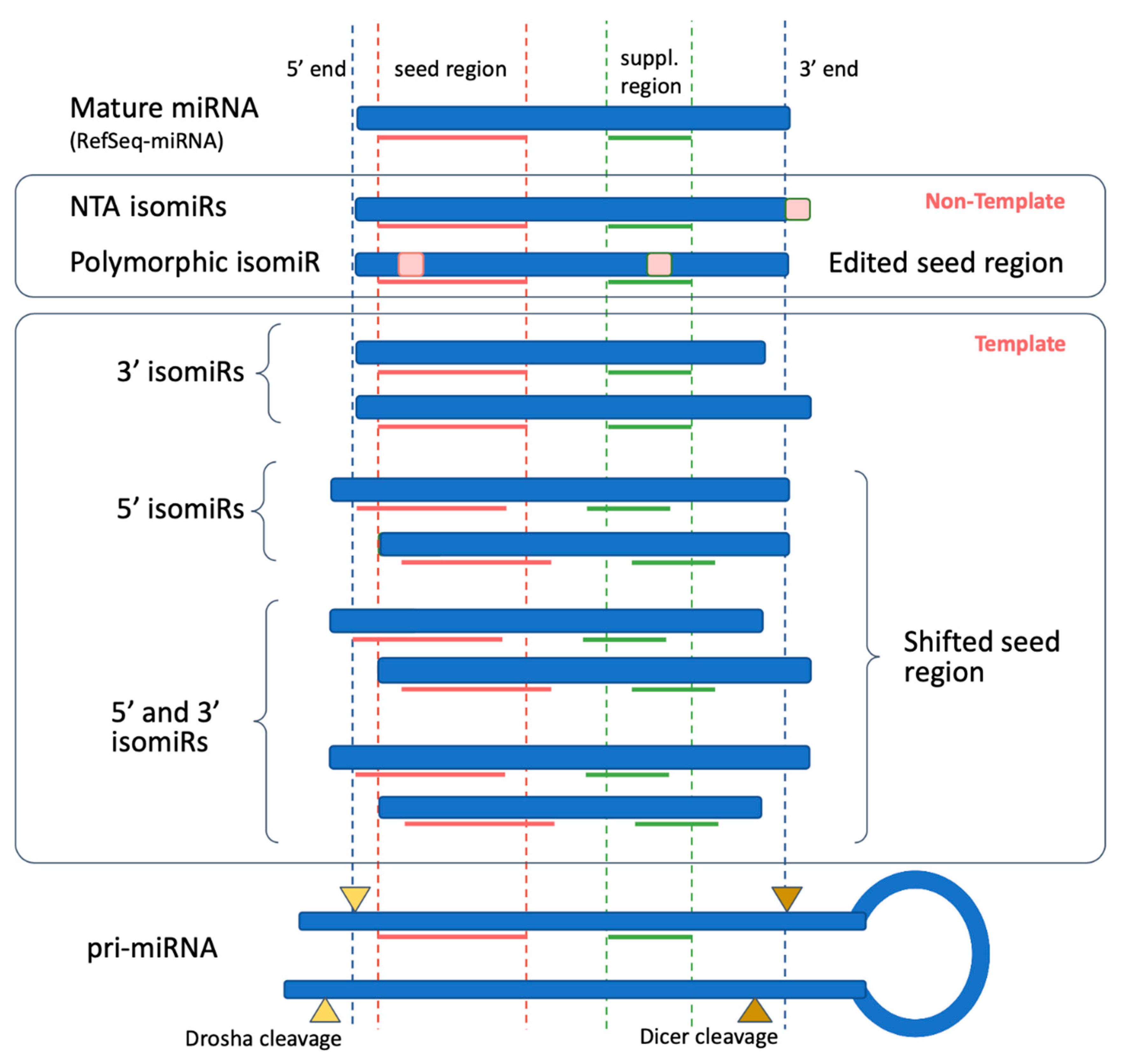



Biomolecules Free Full Text Isomirs Hidden Soldiers In The Mirna Regulatory Army And How To Find Them Html




The Biochemical Basis Of Microrna Targeting Efficacy Science




Mirna Sequencing Report




Frontiers In Bioscience 17 2508 2540 June 1 12 Micrornas Molecular Features And Role In Cancer Elodie Lages1 2 Helene Ipas1 2 Audrey Guttin1 2 3 Houssam Nesr1 2 Francois Berger1 2 Jean Paul Issartel1 2 3 4 1inserm U6 Team7




Clustering Pattern And Functional Effect Of Snps In Human Mirna Seed Regions




Dbmts A Comprehensive Database Of Putative Human Microrna Target Site Snvs And Their Functional Predictions Li Human Mutation Wiley Online Library




A Study Of Micrornas In Silico And In Vivo Bioimaging Of Microrna Biogenesis And Regulation Kim 09 The Febs Journal Wiley Online Library



Mir0c Microrna 0c




Microrna Regulation And Cardiac Calcium Signaling Circulation Research




Dbmts A Comprehensive Database Of Putative Human Microrna Target Site Snvs And Their Functional Predictions Li Human Mutation Wiley Online Library




3 Uridylation Confers Mirnas With Non Canonical Target Repertoires Sciencedirect




A Microrna Guide For Clinicians And Basic Scientists Background And Experimental Techniques Heart Lung And Circulation



Www Longdom Org Open Access Micrornas In Skin Biology Biogenesis Regulations And Functions In Homeostasis And Diseases Pdf




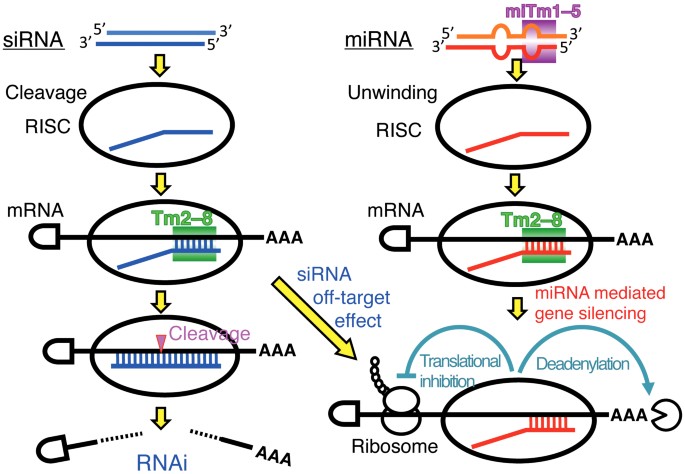
Sirna Versus Mirna As Therapeutics For Gene Silencing Molecular Therapy Nucleic Acids
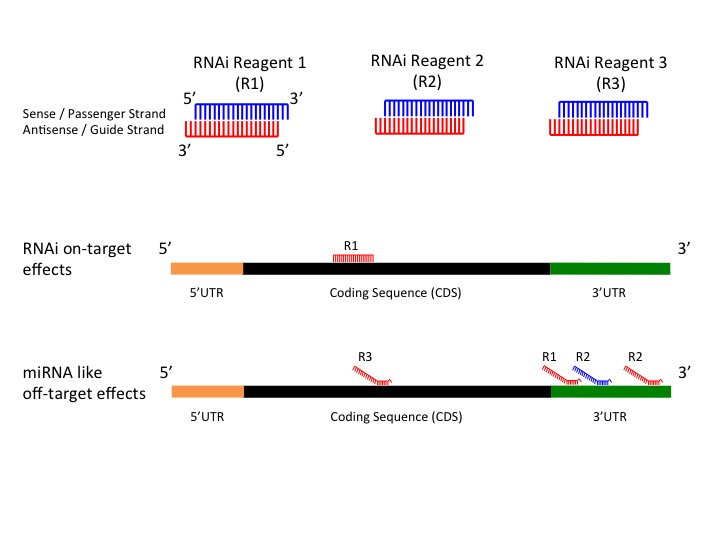



Gess




The Characterization Of Microrna Mediated Gene Regulation As Impacted By Both Target Site Location And Seed Match Type
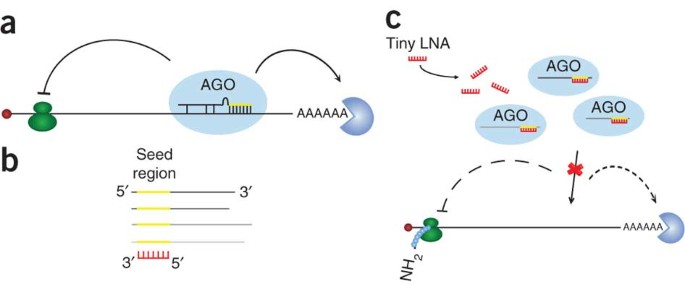



Silencing Of Microrna Families By Seed Targeting Tiny Lnas Nature Genetics




Miraw A Deep Learning Approach To Predict Mirna Targets By Analyzing Whole Mirna Transcripts Biorxiv
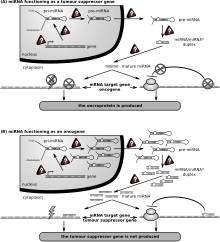



Microrna Wikipedia




The Biochemical Basis Of Microrna Targeting Efficacy Science
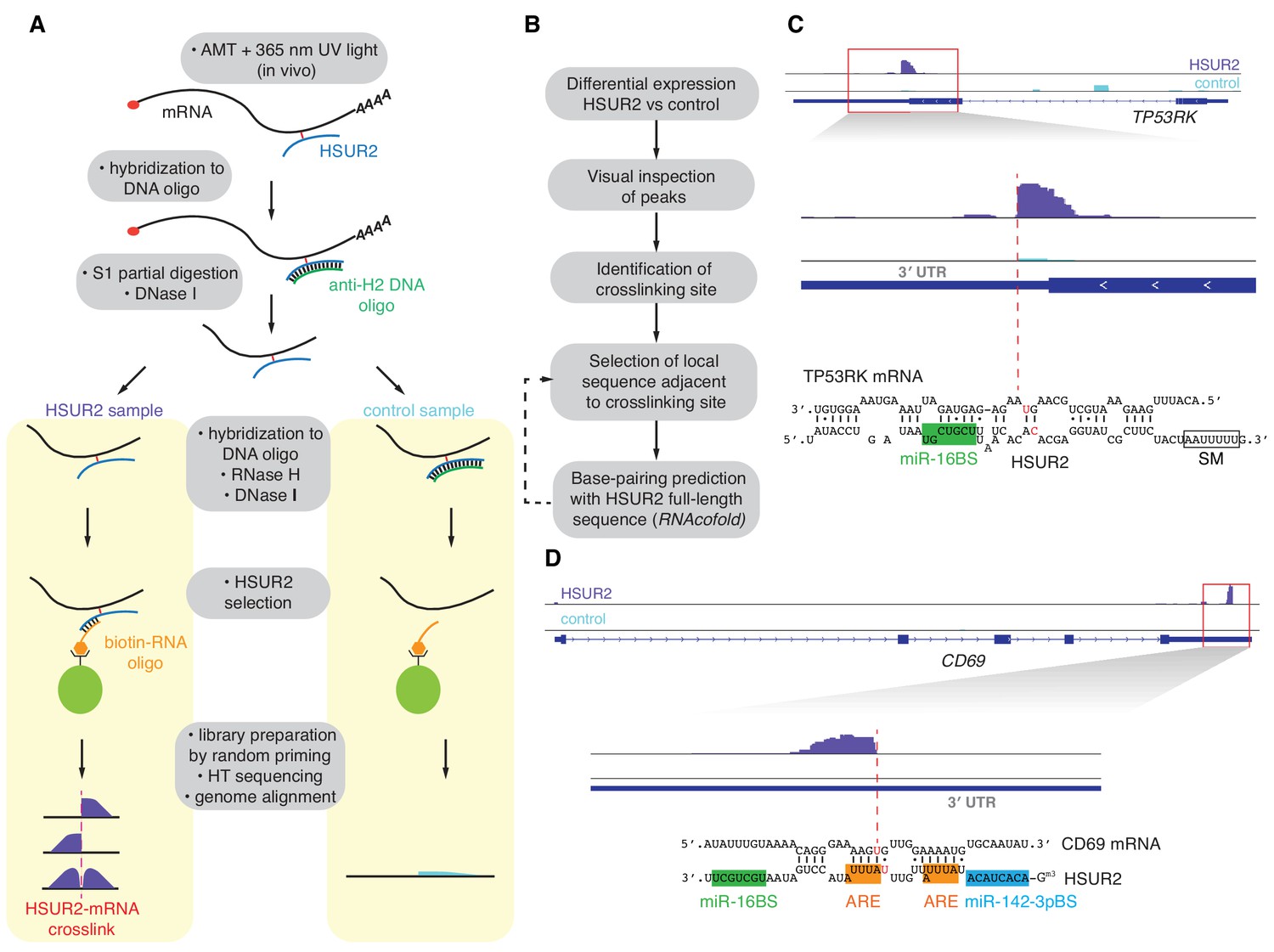



Viral Mirna Adaptor Differentially Recruits Mirnas To Target Mrnas Through Alternative Base Pairing Elife
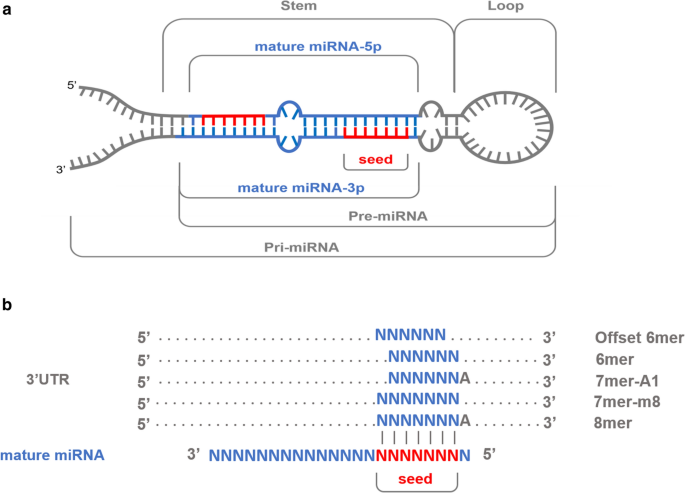



Variability In Porcine Microrna Genes And Its Association With Mrna Expression And Lipid Phenotypes Genetics Selection Evolution Full Text




Human Polymorphism At Micrornas And Microrna Target Sites Pnas
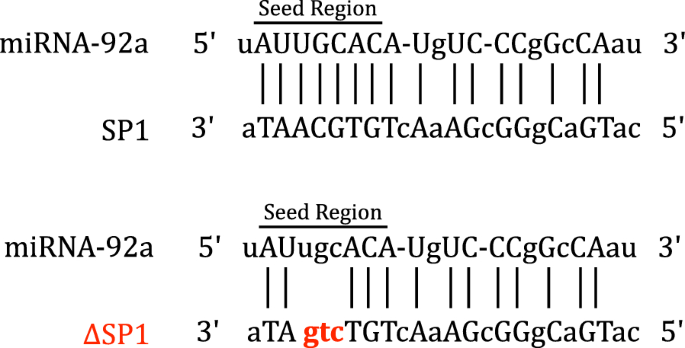



Microrna 92a Regulates The Expression Of Aphid Bacteriocyte Specific Secreted Protein 1 Bmc Research Notes Full Text




Beyond The Seed Structural Basis For Supplementary Microrna Targeting By Human Argonaute2 The Embo Journal




Miraw A Deep Learning Approach To Predict Mirna Targets By Analyzing Whole Mirna Transcripts Biorxiv




Mirna Mirna Targets Functional Analysis




Chemical Modifications In The Seed Region Of Mirnas 221 222 Increase The Silencing Performances In Gastrointestinal Stromal Tumor Cells Sciencedirect



0 件のコメント:
コメントを投稿